class: center, middle, inverse, title-slide # Interactions (II) --- ## Last time... Introduction to interactions with one categorical and one continuous predictor ## Today Two continuous predictors --- ## What are interactions? When we have two variables, A and B, in a regression model, we are testing whether these variables have **additive effects** on our outcome, Y. That is, the effect of A on Y is constant over all values of B. - Example: Drinking coffee and hours of sleep have additive effects on alertness; no matter how any hours I slept the previous night, drinking one cup of coffee will make me .5 SD more awake than not drinking coffee. --- ## What are interactions? However, we may hypothesis that two variables have **joint effects**, or interact with each other. In this case, the effect of A on Y changes as a function of B. - Example: Chronic stress has a negative impact on health but only for individuals who receive little or no social support; for individuals with high social support, chronic stress has no impact on health. - This is also referred to as **moderation.** - The **"interaction term"** is the regression coefficient that tests this hypothesis. --- .pull-left[ ### Univariate regression <!-- --> ] .pull-right[ ### Multivariate regression <!-- --> ] --- ### Multivariate regression with an interaction <!-- --> --- ### Example Here we have an outcome (`Stress`) that we are interested in predicting from trait `Anxiety` and levels of social `Support`. ```r library(tidyverse) stress.data = read_csv("https://raw.githubusercontent.com/uopsych/psy612/master/data/stress.csv") glimpse(stress.data) ``` ``` ## Rows: 118 ## Columns: 5 ## $ id <dbl> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18… ## $ Anxiety <dbl> 10.18520, 5.58873, 6.58500, 8.95430, 7.59910, 8.15600, 9.0802… ## $ Stress <dbl> 3.19813, 7.00840, 6.17400, 8.69884, 5.26707, 5.12485, 6.85380… ## $ Support <dbl> 6.16020, 8.90690, 10.54330, 11.46050, 5.55160, 7.51170, 8.556… ## $ group <chr> "Tx", "Control", "Tx", "Tx", "Control", "Tx", "Tx", "Control"… ``` ```r psych::describe(stress.data, fast = T) ``` ``` ## vars n mean sd min max range se ## id 1 118 59.50 34.21 1.00 118.00 117.00 3.15 ## Anxiety 2 118 7.61 2.49 0.70 14.64 13.94 0.23 ## Stress 3 118 5.18 1.88 0.62 10.32 9.71 0.17 ## Support 4 118 8.73 3.28 0.02 17.34 17.32 0.30 ## group 5 118 NaN NA Inf -Inf -Inf NA ``` --- ```r imodel = lm(Stress ~ Anxiety*Support, data = stress.data) summary(imodel) ``` ``` ## ## Call: ## lm(formula = Stress ~ Anxiety * Support, data = stress.data) ## ## Residuals: ## Min 1Q Median 3Q Max ## -3.8163 -1.0783 0.0373 0.9200 3.6109 ## ## Coefficients: ## Estimate Std. Error t value Pr(>|t|) ## (Intercept) -2.73966 1.12101 -2.444 0.01606 * ## Anxiety 0.61561 0.13010 4.732 6.44e-06 *** ## Support 0.66697 0.09547 6.986 2.02e-10 *** ## Anxiety:Support -0.04174 0.01309 -3.188 0.00185 ** ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Residual standard error: 1.462 on 114 degrees of freedom ## Multiple R-squared: 0.4084, Adjusted R-squared: 0.3928 ## F-statistic: 26.23 on 3 and 114 DF, p-value: 5.645e-13 ``` `$$\hat{Stress} = -2.74 + 0.62(Anx) + 0.67(Sup) + -0.04(Anx \times Sup)$$` --- ## Conditional effects and simple slopes The regression line estimated in this model is quite difficult to interpret on its own. A good strategy is to decompose the regression equation into **simple slopes**, which are determined by calculating the conditional effects at a specific level of the moderating variable. - Simple slope: the equation for Y on X at differnt levels of Z; but also refers to only the coefficient for X in this equation - Conditional effect: the slope coefficients in the full regression model which can change. These are the lower-order terms associated with a variable. E.g., X has a conditional effect on Y. Which variable is the "predictor" (X) and which is the "moderator" (Z)? --- The conditional nature of these effects is easiest to see by "plugging in" different values for one of your variables. Return to the regression equation estimated in our stress data: `$$\hat{Stress} = -2.74 + 0.62(\text{Anx})+ 0.67(\text{Sup})+ -0.04(\text{Anx} \times \text{Sup})$$` -- **Set Support to 5** $$ `\begin{aligned} \hat{Stress} &= -2.74 + 0.62(\text{Anx})+ 0.67(5)+ -0.04(\text{Anx} \times 5) \\ &= -2.74 + 0.62(\text{Anx})+ 3.35+ -0.2(\text{Anx}) \\ &= 0.61 + 0.42(\text{Anx}) \end{aligned}` $$ -- **Set Support to 10** $$ `\begin{aligned} \hat{Stress} &= -2.74 + 0.62(\text{Anx})+ 0.67(10)+ -0.04(\text{Anx} \times 10) \\ &= -2.74 + 0.62(\text{Anx})+ 6.7+ -0.4(\text{Anx}) \\ &= 3.96 + 0.22(\text{Anx}) \end{aligned}` $$ --- ## Plotting interactions What is this plotting? ```r library(sjPlot) plot_model(imodel, type = "pred", term = "Support") ``` <!-- --> --- ## Plotting interactions What is this plotting? ```r plot_model(imodel, type = "pred", terms = c("Support", "Anxiety[mean]")) ``` 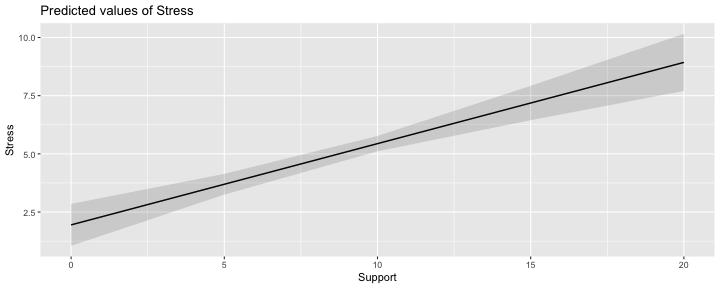<!-- --> --- .pull-left[ 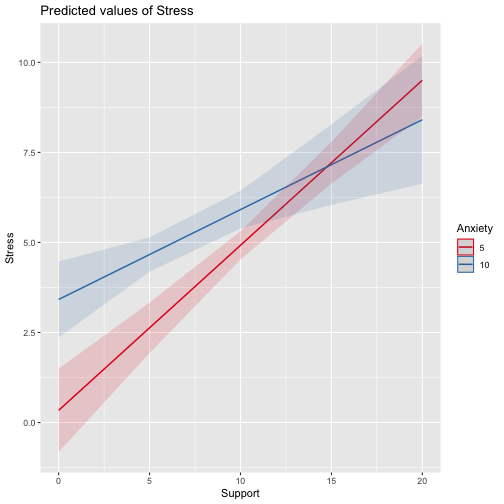<!-- --> ```r plot_model(imodel, type = "pred", terms = c("Support", "Anxiety[5,10]")) ``` Put values of the moderator in brackets! ] .pull-right[ 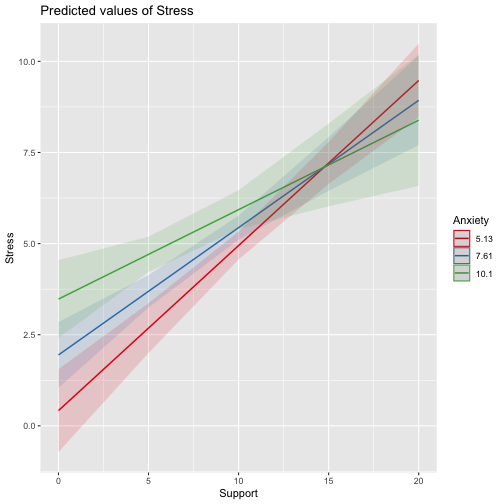<!-- --> ```r plot_model(imodel, type = "pred", terms = c("Support", "Anxiety"), mdrt.values = "meansd") ``` Put values of the moderator in argument `mdrt.values`! ] --- ## Testing simple slopes ```r library(reghelper) anx.vals = list(Anxiety = c(5,6,7)) simple_slopes(imodel, levels = anx.vals) ``` ``` ## Anxiety Support Test Estimate Std. Error t value df Pr(>|t|) Sig. ## 1 5 sstest 0.4583 0.0519 8.8350 114 1.415e-14 *** ## 2 6 sstest 0.4165 0.0493 8.4427 114 1.126e-13 *** ## 3 7 sstest 0.3748 0.0502 7.4652 114 1.797e-11 *** ``` --- ## Testing simple slopes ```r m.anx = mean(stress.data$Anxiety) s.anx = sd(stress.data$Anxiety) anx.vals = list(Anxiety = c(m.anx - s.anx, m.anx, m.anx + s.anx)) simple_slopes(imodel, levels = anx.vals) ``` ``` ## Anxiety Support Test Estimate Std. Error t value df Pr(>|t|) Sig. ## 1 5.12715526773916 sstest 0.4530 0.0514 8.8177 114 1.551e-14 *** ## 2 7.61433127118644 sstest 0.3491 0.0524 6.6656 114 9.824e-10 *** ## 3 10.1015072746337 sstest 0.2453 0.0705 3.4800 114 0.0007112 *** ``` --- ## Simple slopes - Significance tests What if you want to compare slopes to each other? How would we test this? -- The test of the interaction coefficient is equivalent to the test of the difference in slopes at levels of Z separated by 1 unit. ```r coef(summary(imodel)) ``` ``` ## Estimate Std. Error t value Pr(>|t|) ## (Intercept) -2.73966246 1.12100519 -2.443934 1.606052e-02 ## Anxiety 0.61561220 0.13010161 4.731780 6.435373e-06 ## Support 0.66696689 0.09547464 6.985802 2.017698e-10 *## Anxiety:Support -0.04174076 0.01309328 -3.187954 1.849736e-03 ``` --- ## Centering The regression equation built using the raw data is not only diffiuclt to interpret, but often the terms displayed are not relevant to the hypotheses we're interested. - `\(b_0\)` is the expected value when all predictors are 0, but this may never happen in real life - `\(b_1\)` is the slope of X when Z is equal to 0, but this may not ever happen either. **Centering** your variables by subtracting the mean from all values can improve the interpretation of your results. - Remember, a linear transformation does not change associations (correlations) between variables. In this case, it only changes the interpretation for some coefficients --- ### Applying one function to multiple variables ```r stress.c = stress.data %>% mutate( across( c(Anxiety, Support) , ~.x-mean(.x) ) ) glimpse(stress.c) ``` ``` ## Rows: 118 ## Columns: 5 ## $ id <dbl> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18… ## $ Anxiety <dbl> 2.57086873, -2.02560127, -1.02933127, 1.33996873, -0.01523127… ## $ Stress <dbl> 3.19813, 7.00840, 6.17400, 8.69884, 5.26707, 5.12485, 6.85380… ## $ Support <dbl> -2.5697997, 0.1769003, 1.8133003, 2.7305003, -3.1783997, -1.2… ## $ group <chr> "Tx", "Control", "Tx", "Tx", "Control", "Tx", "Tx", "Control"… ``` --- ### Model with centered predictors ```r summary(lm(Stress ~ Anxiety*Support, data = stress.c)) ``` ``` ## ## Call: ## lm(formula = Stress ~ Anxiety * Support, data = stress.c) ## ## Residuals: ## Min 1Q Median 3Q Max ## -3.8163 -1.0783 0.0373 0.9200 3.6109 ## ## Coefficients: ## Estimate Std. Error t value Pr(>|t|) ## (Intercept) 4.99580 0.14647 34.108 < 2e-16 *** ## Anxiety 0.25122 0.06489 3.872 0.000181 *** ## Support 0.34914 0.05238 6.666 9.82e-10 *** ## Anxiety:Support -0.04174 0.01309 -3.188 0.001850 ** ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Residual standard error: 1.462 on 114 degrees of freedom ## Multiple R-squared: 0.4084, Adjusted R-squared: 0.3928 ## F-statistic: 26.23 on 3 and 114 DF, p-value: 5.645e-13 ``` --- ### Model with uncentered predictors ```r summary(lm(Stress ~ Anxiety*Support, data = stress.data)) ``` ``` ## ## Call: ## lm(formula = Stress ~ Anxiety * Support, data = stress.data) ## ## Residuals: ## Min 1Q Median 3Q Max ## -3.8163 -1.0783 0.0373 0.9200 3.6109 ## ## Coefficients: ## Estimate Std. Error t value Pr(>|t|) ## (Intercept) -2.73966 1.12101 -2.444 0.01606 * ## Anxiety 0.61561 0.13010 4.732 6.44e-06 *** ## Support 0.66697 0.09547 6.986 2.02e-10 *** ## Anxiety:Support -0.04174 0.01309 -3.188 0.00185 ** ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Residual standard error: 1.462 on 114 degrees of freedom ## Multiple R-squared: 0.4084, Adjusted R-squared: 0.3928 ## F-statistic: 26.23 on 3 and 114 DF, p-value: 5.645e-13 ``` --- ## Standardized regression equation If you're interested in getting the standardized regression equation, you can follow the procedure of standardizing your variables first and then entering them into your linear model. An important note: You must take the product of the Z-scores, not the Z-score of the products to get the correct regression model. .pull-left[ #### This is OK ```r z(Y) ~ z(X) + z(Z) + z(X)*z(Z) z(Y) ~ z(X)*z(Z) ``` ] .pull-right[ #### This is not OK ```r z(Y) ~ z(X) + z(Z) + z(X*Z) ``` ] --- ### Applying one function to all numeric variables ```r stress.z = stress.data %>% mutate(across(where(is.numeric), scale)) head(stress.z) ``` ``` ## # A tibble: 6 x 5 ## id[,1] Anxiety[,1] Stress[,1] Support[,1] group ## <dbl> <dbl> <dbl> <dbl> <chr> ## 1 -1.71 1.03 -1.06 -0.784 Tx ## 2 -1.68 -0.814 0.975 0.0540 Control ## 3 -1.65 -0.414 0.530 0.553 Tx ## 4 -1.62 0.539 1.88 0.833 Tx ## 5 -1.59 -0.00612 0.0464 -0.970 Control ## 6 -1.56 0.218 -0.0294 -0.372 Tx ``` --- ### Standardized equation ```r summary(lm(Stress ~ Anxiety*Support, stress.z)) ``` ``` ## ## Call: ## lm(formula = Stress ~ Anxiety * Support, data = stress.z) ## ## Residuals: ## Min 1Q Median 3Q Max ## -2.03400 -0.57471 0.01989 0.49037 1.92453 ## ## Coefficients: ## Estimate Std. Error t value Pr(>|t|) ## (Intercept) -0.09818 0.07807 -1.258 0.211093 ## Anxiety 0.33302 0.08602 3.872 0.000181 *** ## Support 0.60987 0.09149 6.666 9.82e-10 *** ## Anxiety:Support -0.18134 0.05688 -3.188 0.001850 ** ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Residual standard error: 0.7792 on 114 degrees of freedom ## Multiple R-squared: 0.4084, Adjusted R-squared: 0.3928 ## F-statistic: 26.23 on 3 and 114 DF, p-value: 5.645e-13 ``` --- ### Unstandardized equation ```r summary(lm(Stress ~ Anxiety*Support, stress.data)) ``` ``` ## ## Call: ## lm(formula = Stress ~ Anxiety * Support, data = stress.data) ## ## Residuals: ## Min 1Q Median 3Q Max ## -3.8163 -1.0783 0.0373 0.9200 3.6109 ## ## Coefficients: ## Estimate Std. Error t value Pr(>|t|) ## (Intercept) -2.73966 1.12101 -2.444 0.01606 * ## Anxiety 0.61561 0.13010 4.732 6.44e-06 *** ## Support 0.66697 0.09547 6.986 2.02e-10 *** ## Anxiety:Support -0.04174 0.01309 -3.188 0.00185 ** ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Residual standard error: 1.462 on 114 degrees of freedom ## Multiple R-squared: 0.4084, Adjusted R-squared: 0.3928 ## F-statistic: 26.23 on 3 and 114 DF, p-value: 5.645e-13 ``` --- # Interactions with two categorical variables If both X and Z are categorical variables, the interpretation of coefficients is no longer the value of means and slopes, but means and differences in means. --- ## Example .pull-left[  ] .pull-right[ Recall our hand-eye coordination study (for a reminder of the study design, see the [ANOVA lecture](https://uopsych.github.io/psy612/lectures/10-anova.html#17)): ] ```r handeye_d = read.csv("https://raw.githubusercontent.com/uopsych/psy612/master/data/hand_eye_task.csv") glimpse(handeye_d) ``` ``` ## Rows: 180 ## Columns: 4 ## $ X <int> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, … ## $ Time <dbl> 623.1855, 547.8381, 709.5921, 815.2066, 719.5926, 732.8989, 566… ## $ Speed <chr> "Slow", "Slow", "Slow", "Slow", "Slow", "Slow", "Slow", "Slow",… ## $ Noise <chr> "None", "None", "None", "None", "None", "None", "None", "None",… ``` --- ```r handeye_d %>% group_by(Speed, Noise) %>% count() ``` ``` ## # A tibble: 9 x 3 ## # Groups: Speed, Noise [9] ## Speed Noise n ## <chr> <chr> <int> ## 1 Fast Controllable 20 ## 2 Fast None 20 ## 3 Fast Uncontrollable 20 ## 4 Medium Controllable 20 ## 5 Medium None 20 ## 6 Medium Uncontrollable 20 ## 7 Slow Controllable 20 ## 8 Slow None 20 ## 9 Slow Uncontrollable 20 ``` --- ### Model summary ```r handeye_d.mod = lm(Time ~ Speed*Noise, data = handeye_d) summary(handeye_d.mod) ``` ``` ## ## Call: ## lm(formula = Time ~ Speed * Noise, data = handeye_d) ## ## Residuals: ## Min 1Q Median 3Q Max ## -316.23 -70.82 4.99 79.87 244.40 ## ## Coefficients: ## Estimate Std. Error t value Pr(>|t|) *## (Intercept) 287.234 25.322 11.343 < 2e-16 *** ## SpeedMedium 205.483 35.811 5.738 4.27e-08 *** ## SpeedSlow 289.439 35.811 8.082 1.11e-13 *** ## NoiseNone 42.045 35.811 1.174 0.24200 ## NoiseUncontrollable -19.072 35.811 -0.533 0.59502 ## SpeedMedium:NoiseNone -9.472 50.644 -0.187 0.85185 ## SpeedSlow:NoiseNone 12.007 50.644 0.237 0.81287 ## SpeedMedium:NoiseUncontrollable -169.023 50.644 -3.337 0.00104 ** ## SpeedSlow:NoiseUncontrollable 36.843 50.644 0.727 0.46792 ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Residual standard error: 113.2 on 171 degrees of freedom ## Multiple R-squared: 0.6109, Adjusted R-squared: 0.5927 ## F-statistic: 33.56 on 8 and 171 DF, p-value: < 2.2e-16 ``` --- ### Model summary ```r handeye_d.mod = lm(Time ~ Speed*Noise, data = handeye_d) summary(handeye_d.mod) ``` ``` ## ## Call: ## lm(formula = Time ~ Speed * Noise, data = handeye_d) ## ## Residuals: ## Min 1Q Median 3Q Max ## -316.23 -70.82 4.99 79.87 244.40 ## ## Coefficients: ## Estimate Std. Error t value Pr(>|t|) ## (Intercept) 287.234 25.322 11.343 < 2e-16 *** *## SpeedMedium 205.483 35.811 5.738 4.27e-08 *** *## SpeedSlow 289.439 35.811 8.082 1.11e-13 *** ## NoiseNone 42.045 35.811 1.174 0.24200 ## NoiseUncontrollable -19.072 35.811 -0.533 0.59502 ## SpeedMedium:NoiseNone -9.472 50.644 -0.187 0.85185 ## SpeedSlow:NoiseNone 12.007 50.644 0.237 0.81287 ## SpeedMedium:NoiseUncontrollable -169.023 50.644 -3.337 0.00104 ** ## SpeedSlow:NoiseUncontrollable 36.843 50.644 0.727 0.46792 ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Residual standard error: 113.2 on 171 degrees of freedom ## Multiple R-squared: 0.6109, Adjusted R-squared: 0.5927 ## F-statistic: 33.56 on 8 and 171 DF, p-value: < 2.2e-16 ``` --- ### Model summary ```r handeye_d.mod = lm(Time ~ Speed*Noise, data = handeye_d) summary(handeye_d.mod) ``` ``` ## ## Call: ## lm(formula = Time ~ Speed * Noise, data = handeye_d) ## ## Residuals: ## Min 1Q Median 3Q Max ## -316.23 -70.82 4.99 79.87 244.40 ## ## Coefficients: ## Estimate Std. Error t value Pr(>|t|) ## (Intercept) 287.234 25.322 11.343 < 2e-16 *** ## SpeedMedium 205.483 35.811 5.738 4.27e-08 *** ## SpeedSlow 289.439 35.811 8.082 1.11e-13 *** *## NoiseNone 42.045 35.811 1.174 0.24200 *## NoiseUncontrollable -19.072 35.811 -0.533 0.59502 ## SpeedMedium:NoiseNone -9.472 50.644 -0.187 0.85185 ## SpeedSlow:NoiseNone 12.007 50.644 0.237 0.81287 ## SpeedMedium:NoiseUncontrollable -169.023 50.644 -3.337 0.00104 ** ## SpeedSlow:NoiseUncontrollable 36.843 50.644 0.727 0.46792 ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Residual standard error: 113.2 on 171 degrees of freedom ## Multiple R-squared: 0.6109, Adjusted R-squared: 0.5927 ## F-statistic: 33.56 on 8 and 171 DF, p-value: < 2.2e-16 ``` --- ### Model summary ```r handeye_d.mod = lm(Time ~ Speed*Noise, data = handeye_d) summary(handeye_d.mod) ``` ``` ## ## Call: ## lm(formula = Time ~ Speed * Noise, data = handeye_d) ## ## Residuals: ## Min 1Q Median 3Q Max ## -316.23 -70.82 4.99 79.87 244.40 ## ## Coefficients: ## Estimate Std. Error t value Pr(>|t|) ## (Intercept) 287.234 25.322 11.343 < 2e-16 *** ## SpeedMedium 205.483 35.811 5.738 4.27e-08 *** ## SpeedSlow 289.439 35.811 8.082 1.11e-13 *** ## NoiseNone 42.045 35.811 1.174 0.24200 ## NoiseUncontrollable -19.072 35.811 -0.533 0.59502 *## SpeedMedium:NoiseNone -9.472 50.644 -0.187 0.85185 *## SpeedSlow:NoiseNone 12.007 50.644 0.237 0.81287 *## SpeedMedium:NoiseUncontrollable -169.023 50.644 -3.337 0.00104 ** *## SpeedSlow:NoiseUncontrollable 36.843 50.644 0.727 0.46792 ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Residual standard error: 113.2 on 171 degrees of freedom ## Multiple R-squared: 0.6109, Adjusted R-squared: 0.5927 ## F-statistic: 33.56 on 8 and 171 DF, p-value: < 2.2e-16 ``` --- ### Model summary ```r handeye_d.mod = lm(Time ~ Speed*Noise, data = handeye_d) summary(handeye_d.mod) ``` ``` ## ## Call: ## lm(formula = Time ~ Speed * Noise, data = handeye_d) ## ## Residuals: ## Min 1Q Median 3Q Max ## -316.23 -70.82 4.99 79.87 244.40 ## ## Coefficients: ## Estimate Std. Error t value Pr(>|t|) ## (Intercept) 287.234 25.322 11.343 < 2e-16 *** ## SpeedMedium 205.483 35.811 5.738 4.27e-08 *** ## SpeedSlow 289.439 35.811 8.082 1.11e-13 *** ## NoiseNone 42.045 35.811 1.174 0.24200 ## NoiseUncontrollable -19.072 35.811 -0.533 0.59502 ## SpeedMedium:NoiseNone -9.472 50.644 -0.187 0.85185 ## SpeedSlow:NoiseNone 12.007 50.644 0.237 0.81287 ## SpeedMedium:NoiseUncontrollable -169.023 50.644 -3.337 0.00104 ** ## SpeedSlow:NoiseUncontrollable 36.843 50.644 0.727 0.46792 ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Residual standard error: 113.2 on 171 degrees of freedom *## Multiple R-squared: 0.6109, Adjusted R-squared: 0.5927 ## F-statistic: 33.56 on 8 and 171 DF, p-value: < 2.2e-16 ``` --- ### Plotting results ```r plot_model(handeye_d.mod, type = "pred", terms = c("Speed", "Noise")) ``` 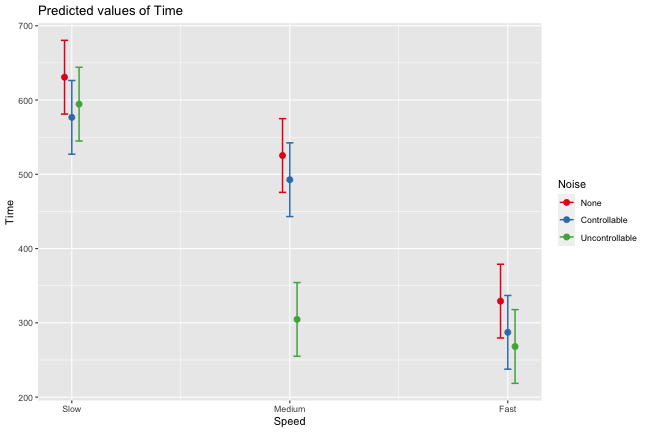<!-- --> --- Remember, regression and ANOVA are mathematically equivalent -- both divide the total variability in `\(Y\)` into variability overlapping with ("explained by") the model and residual variability. What differs is the way results are presented. The regression framework is excellent for continuous variables, but interpreting the interactions of categorical variables is more difficult. So we'll switch back to the ANOVA framework next time and talk about... --- class: inverse ## Next time Factorial ANOVA