Bonus lecture
Last time
Paired-samples t-tests
- aka one-sample t-tests on difference scores
Today
- Repeated measures
- Intro to text analysis
- Bootstrapping?
Repeated Measures
- How does the number of trials per participant affect statistical power?
- Can more trials compensate for fewer participants?
- How do we analyze experiments with multiple trials per participant?
Key Concepts
- In cognitive experiments, we have:
- Multiple participants (n)
- Multiple trials per participant (k)
- Two or more conditions
- Each trial produces a measurement
- Multiple trials can:
- Improve measurement precision
- Reduce within-subject variability
- Increase reliability
Simulation: Effect of Trials
(Function to simulate experiment here)
Code
simulate_cognitive_experiment <- function(
n_participants = 20, # number of participants
n_trials = 50, # trials per condition per participant
true_effect = 0.5, # mean difference between conditions
participant_sd = 0.25, # between-participant variability
trial_sd = 1.0, # within-participant (trial) variability
seed = 123 # for reproducibility
) {
set.seed(seed)
# Generate participant random effects (individual differences)
participant_effects <- rnorm(n_participants, mean = 0, sd = participant_sd)
# Create a data frame with all combinations of participants and trials
experiment_data <- expand_grid(
participant = 1:n_participants,
trial = 1:n_trials,
condition = c("A", "B")
) %>%
# Add participant-level random effects
mutate(
participant_effect = rep(participant_effects, each = n_trials * 2),
# Add condition effect (only for condition B)
condition_effect = if_else(condition == "B", true_effect, 0),
# Generate trial-level noise
trial_noise = rnorm(n(), mean = 0, sd = trial_sd),
# Compute response time (or other DV)
response = 0.5 + participant_effect + condition_effect + trial_noise
) %>%
# Reorder columns for clarity
select(participant, condition, trial, response)
return(experiment_data)
}# Generate example data
data <- simulate_cognitive_experiment(
n_participants = 20,
n_trials = 100,
true_effect = 0.5,
participant_sd = 0.25,
trial_sd = 1.0
)
# View first few rows
data# A tibble: 4,000 × 4
participant condition trial response
<int> <chr> <int> <dbl>
1 1 A 1 -0.708
2 1 B 1 0.642
3 1 A 2 -0.666
4 1 B 2 0.131
5 1 A 3 -0.265
6 1 B 3 -0.827
7 1 A 4 1.20
8 1 B 4 1.01
9 1 A 5 -0.778
10 1 B 5 2.11
# ℹ 3,990 more rows# Participant-level summary
data %>%
group_by(participant, condition) %>%
summarise(
mean_rt = mean(response),
sd_rt = sd(response),
n_trials = n(),
.groups = "drop"
) # A tibble: 40 × 5
participant condition mean_rt sd_rt n_trials
<int> <chr> <dbl> <dbl> <int>
1 1 A 0.345 0.935 100
2 1 B 0.842 0.967 100
3 2 A 0.562 0.929 100
4 2 B 0.853 1.03 100
5 3 A 1.07 1.04 100
6 3 B 1.27 0.914 100
7 4 A 0.558 1.01 100
8 4 B 0.991 1.03 100
9 5 A 0.561 1.02 100
10 5 B 1.12 1.14 100
# ℹ 30 more rows# A tibble: 20 × 4
participant A B effect_size
<int> <dbl> <dbl> <dbl>
1 1 0.345 0.842 0.497
2 2 0.562 0.853 0.291
3 3 1.07 1.27 0.200
4 4 0.558 0.991 0.433
5 5 0.561 1.12 0.564
6 6 0.977 1.38 0.401
7 7 0.597 1.15 0.555
8 8 0.139 0.881 0.742
9 9 0.315 0.911 0.596
10 10 0.395 1.01 0.620
11 11 0.752 1.17 0.416
12 12 0.543 0.947 0.405
13 13 0.662 1.17 0.510
14 14 0.488 0.975 0.486
15 15 0.480 0.927 0.447
16 16 0.845 1.38 0.534
17 17 0.562 1.20 0.634
18 18 -0.107 0.688 0.796
19 19 0.582 0.992 0.410
20 20 0.561 0.940 0.379# A tibble: 20 × 4
participant A B effect_size
<int> <dbl> <dbl> <dbl>
1 1 0.238 1.04 0.800
2 2 0.627 0.696 0.0686
3 3 1.15 1.30 0.158
4 4 0.324 1.05 0.721
5 5 0.524 1.11 0.583
6 6 1.05 1.58 0.529
7 7 0.590 1.21 0.616
8 8 0.0702 0.980 0.910
9 9 0.127 1.07 0.947
10 10 0.321 0.900 0.579
11 11 0.904 1.40 0.495
12 12 0.413 0.736 0.322
13 13 0.507 1.24 0.729
14 14 0.901 0.945 0.0440
15 15 0.401 1.12 0.715
16 16 0.812 1.14 0.332
17 17 0.568 1.38 0.809
18 18 -0.146 0.841 0.987
19 19 0.610 1.15 0.538
20 20 0.955 0.977 0.0217Why More Trials Help
Law of Large Numbers
- Individual trial measurements are noisy
- Mean of many trials is more stable
- Reduces measurement error
Standard Error Formula
- SE = σ / √n
- More trials → smaller SE of participant means
- Smaller SE → More power
Code
participant_effects %>%
mutate(Trials = 100) %>%
full_join(
mutate(participant_effects20, Trials = 20)
) %>%
pivot_longer(cols = c("A", "B"),
names_to = "condition",
values_to = "avg_response") %>%
ggplot(., aes(x = condition,
y = avg_response,
fill = as.factor(Trials))) +
geom_boxplot() +
labs(title = "Reduced Variability with More Trials",
y = "Mean Score",
fill = "Number of Trials") +
theme_minimal() +
theme(legend.position = "bottom")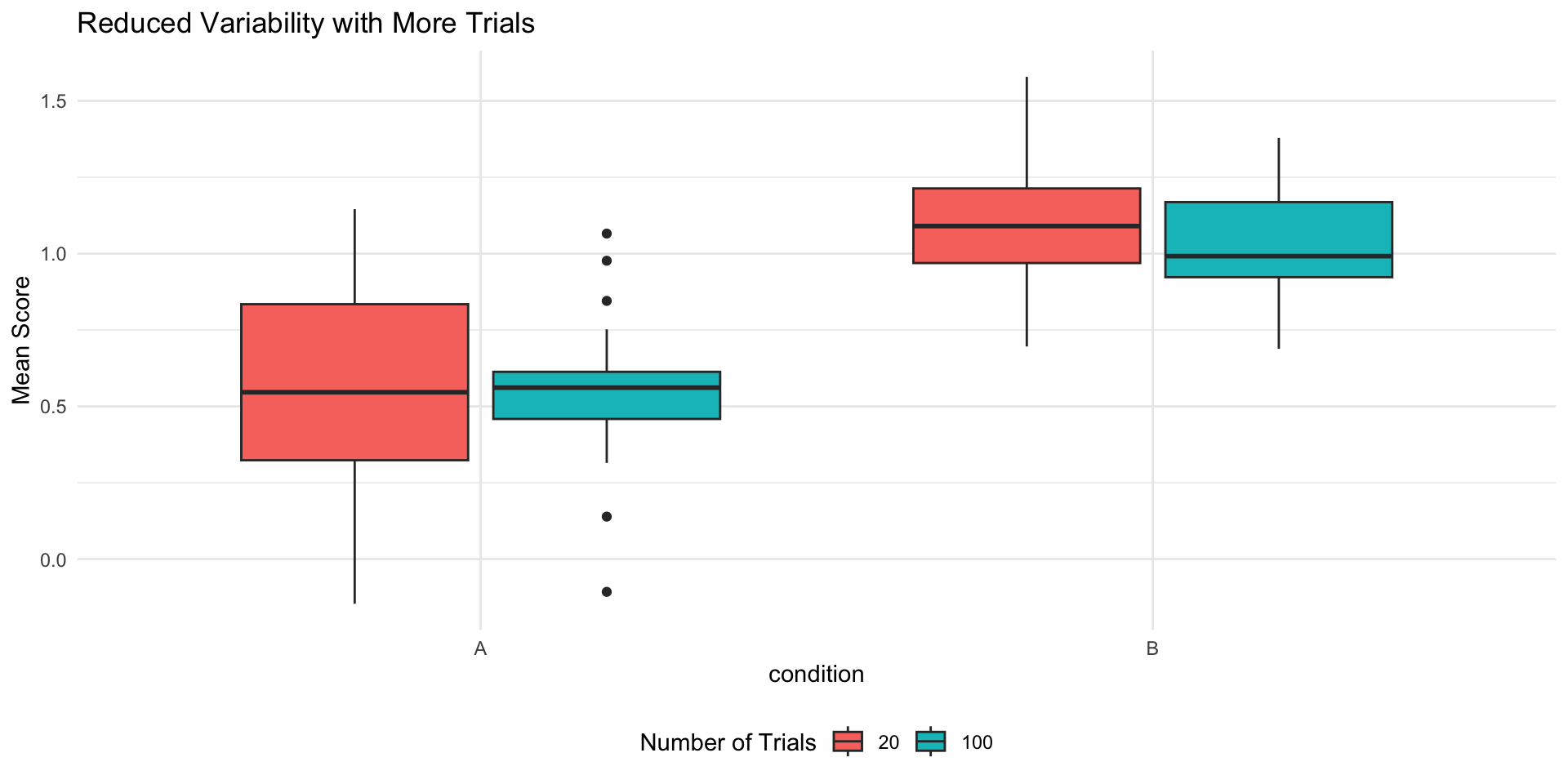
Practical Guidelines
- More trials generally help, but with diminishing returns
- Consider:
- Participant fatigue
- Practice effects
- Time constraints
- Resource limitations
Analysis Approaches
Summary by Dan McNeish
- Participant-level Analysis
- Average trials for each participant
- Run paired t-test on participant means
- Simple but may lose information
- Multilevel modeling
- Use all information
- Especially useful when interested in individual differences
- Clustered errors and GEE
- Don’t care about individual differences
- Fixed effects models
- Useful when relevant contextual factors are not measured
Coming Soon (PSY 613)…
- Mixed-effects models
- Account for trial-level variation
- Handle missing data
- Model individual differences
- Power analysis for repeated measures
Linear mixed model fit by REML ['lmerMod']
Formula: response ~ 1 + condition + (1 | participant)
Data: data
REML criterion at convergence: 11358.8
Scaled residuals:
Min 1Q Median 3Q Max
-3.2348 -0.6633 -0.0035 0.6758 3.4111
Random effects:
Groups Name Variance Std.Dev.
participant (Intercept) 0.0416 0.2039
Residual 0.9888 0.9944
Number of obs: 4000, groups: participant, 20
Fixed effects:
Estimate Std. Error t value
(Intercept) 0.54415 0.05074 10.72
conditionB 0.49573 0.03145 15.77
Correlation of Fixed Effects:
(Intr)
conditionB -0.310Text analysis
Data came from a study of parents of young children, collected during 2020.
Parents answered the question: “How do you feel about the COVID-19 vaccine in terms of its safety and effectiveness, and what are your plans in terms of whether or not to get it?”
Data Preparation
Let’s examine and clean our text data:
Rows: 6,554
Columns: 7
$ ...1 <dbl> 23435, 23436, 23437, 23439, 23440, 23444, 23445, 23451, 2…
$ CaregiverID <chr> "R3ID00020", "R3ID00032", "R3ID00032", "R3ID00036", "R3ID…
$ StartDate <dttm> 2021-06-11 08:15:52, 2021-06-09 10:59:01, 2021-11-10 09:…
$ CaregiverAge <lgl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
$ DEMO.006.a <lgl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
$ DEMO.014 <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
$ HEALTH.030 <chr> "I am not planning on it right now but i am open to the f…Cleaning text requires the use of “regular expressions.” This is like a sub-dialect of coding. The website regex101.com is very useful for navigating this code, but of course, AI is great too.
# Create a clean text column
rapid_clean <- rapid %>%
mutate(
response = HEALTH.030,
# Convert to lowercase
response = tolower(response),
# Remove punctuation
response = str_remove_all(response, "\\?"),
response = str_replace_all(response, "[[:punct:]]", " "),
# Remove extra whitespace
response = str_squish(response)
)
head(rapid_clean$response) # show first few responses[1] "i am not planning on it right now but i am open to the future"
[2] "i dont feel it is safe yet or ready"
[3] "im not sure about the vaccine and its effectiveness im still weary and havent decided on taking the vaccine yet"
[4] "i dont feel like it works i had to get it for work"
[5] "i already got both doses"
[6] "i got it" Breaking Text into Words
We’ll use tidytext to tokenize our responses:
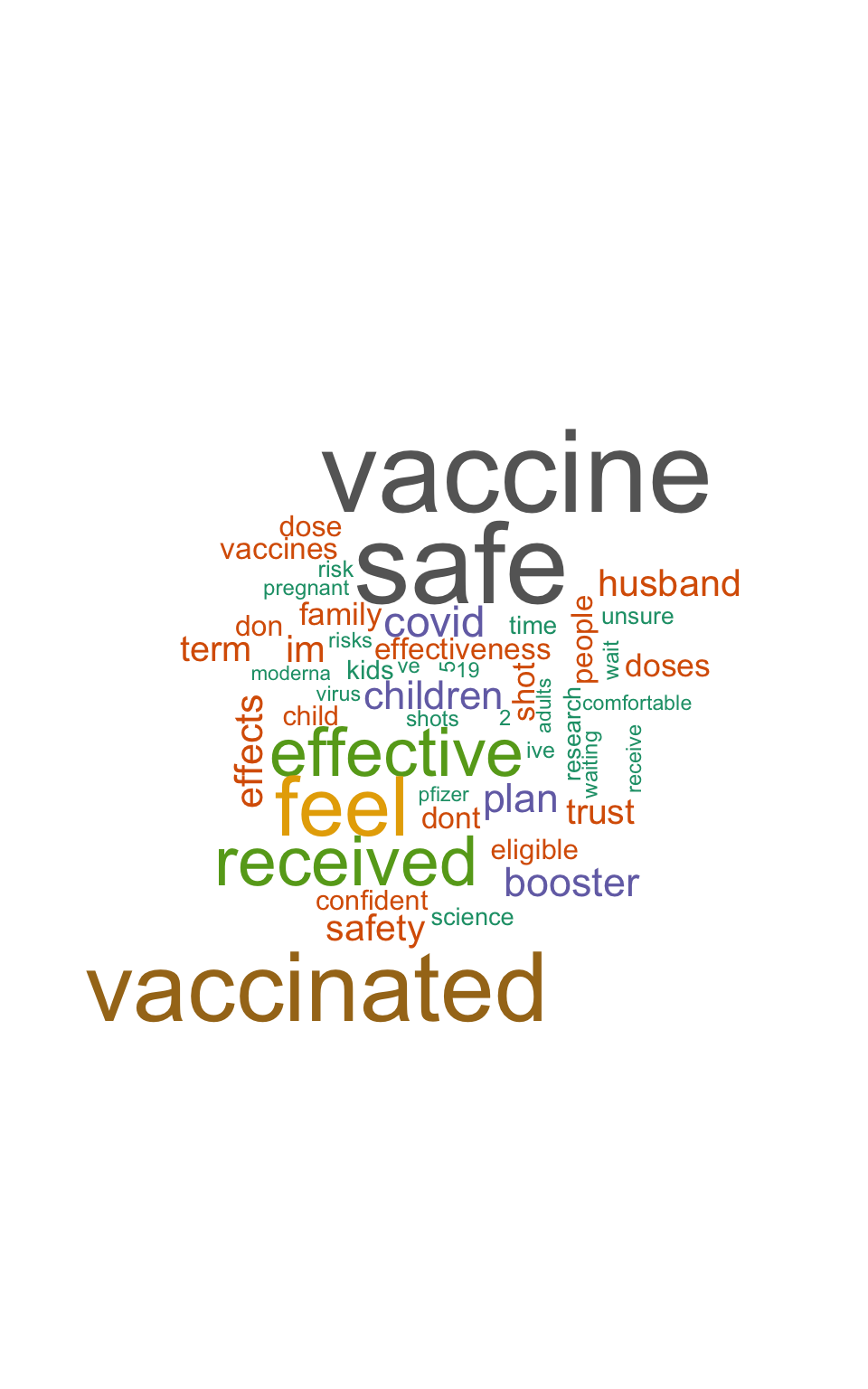
| word | n |
|---|---|
| vaccine | 1789 |
| safe | 1747 |
| vaccinated | 1457 |
| feel | 1210 |
| received | 976 |
| effective | 952 |
| covid | 514 |
| booster | 463 |
| plan | 460 |
| children | 449 |
Visualizing Common Words
Let’s create a word cloud:
Finding Themes: Bigrams
Let’s look at word pairs to understand context better:
Code
bigrams_df <- rapid_clean %>%
unnest_tokens(bigram, response, token = "ngrams", n = 2) %>%
separate(bigram, c("word1", "word2"), sep = " ") %>%
filter(!word1 %in% stop_words$word,
!word2 %in% stop_words$word) %>%
filter(!is.na(word1)) %>%
filter(!is.na(word2))
# View top bigrams
bigrams_df %>%
unite(bigram, word1, word2, sep = " ") %>%
count(bigram, sort = TRUE) %>%
slice_head(n = 10) %>%
knitr::kable()| bigram | n |
|---|---|
| term effects | 154 |
| covid 19 | 118 |
| feel confident | 65 |
| 19 vaccine | 47 |
| dont feel | 46 |
| dont trust | 44 |
| covid vaccine | 43 |
| feel comfortable | 43 |
| children vaccinated | 41 |
| feel safe | 39 |
Sentiment Analysis
Let’s analyze the emotional content of responses:
# A tibble: 10 × 2
word sentiment
<chr> <chr>
1 shark negative
2 awsome positive
3 irrepressible negative
4 humiliation negative
5 disgruntled negative
6 preferably positive
7 comforting positive
8 immobilized negative
9 bored negative
10 eye-catching positive Sentiment Analysis
# Add sentiment scores
sentiment_df <- words_df %>%
inner_join(get_sentiments("bing")) %>%
count(CaregiverID, StartDate, sentiment) %>%
pivot_wider(names_from = sentiment,
values_from = n,
values_fill = 0) %>%
mutate(sentiment_score = positive - negative)
sentiment_df# A tibble: 4,070 × 5
CaregiverID StartDate positive negative sentiment_score
<chr> <dttm> <int> <int> <int>
1 R3ID00032 2021-06-09 10:59:01 2 0 2
2 R3ID00032 2021-11-10 09:57:51 1 1 0
3 R3ID00049 2021-12-10 08:17:19 1 0 1
4 R3ID00083 2021-11-14 09:11:26 0 1 -1
5 R3ID00084 2021-06-11 10:58:56 1 0 1
6 R3ID00084 2021-10-13 09:21:12 1 0 1
7 R3ID00117 2021-12-08 10:30:31 0 1 -1
8 R3ID00166 2021-03-17 22:15:04 1 0 1
9 R3ID00168 2021-12-08 10:13:24 1 0 1
10 R3ID00188 2021-10-14 09:17:07 1 0 1
# ℹ 4,060 more rowsSentiment Analysis
Code
# View distribution of sentiment
ggplot(sentiment_df, aes(x = sentiment_score)) +
geom_histogram(binwidth = 1, fill = "steelblue", alpha = 0.7) +
geom_vline(aes(xintercept = 0), linetype = "dashed") +
labs(title = "Distribution of Sentiment Scores",
x = "Sentiment Score (Positive - Negative)",
y = "Count") +
theme_minimal()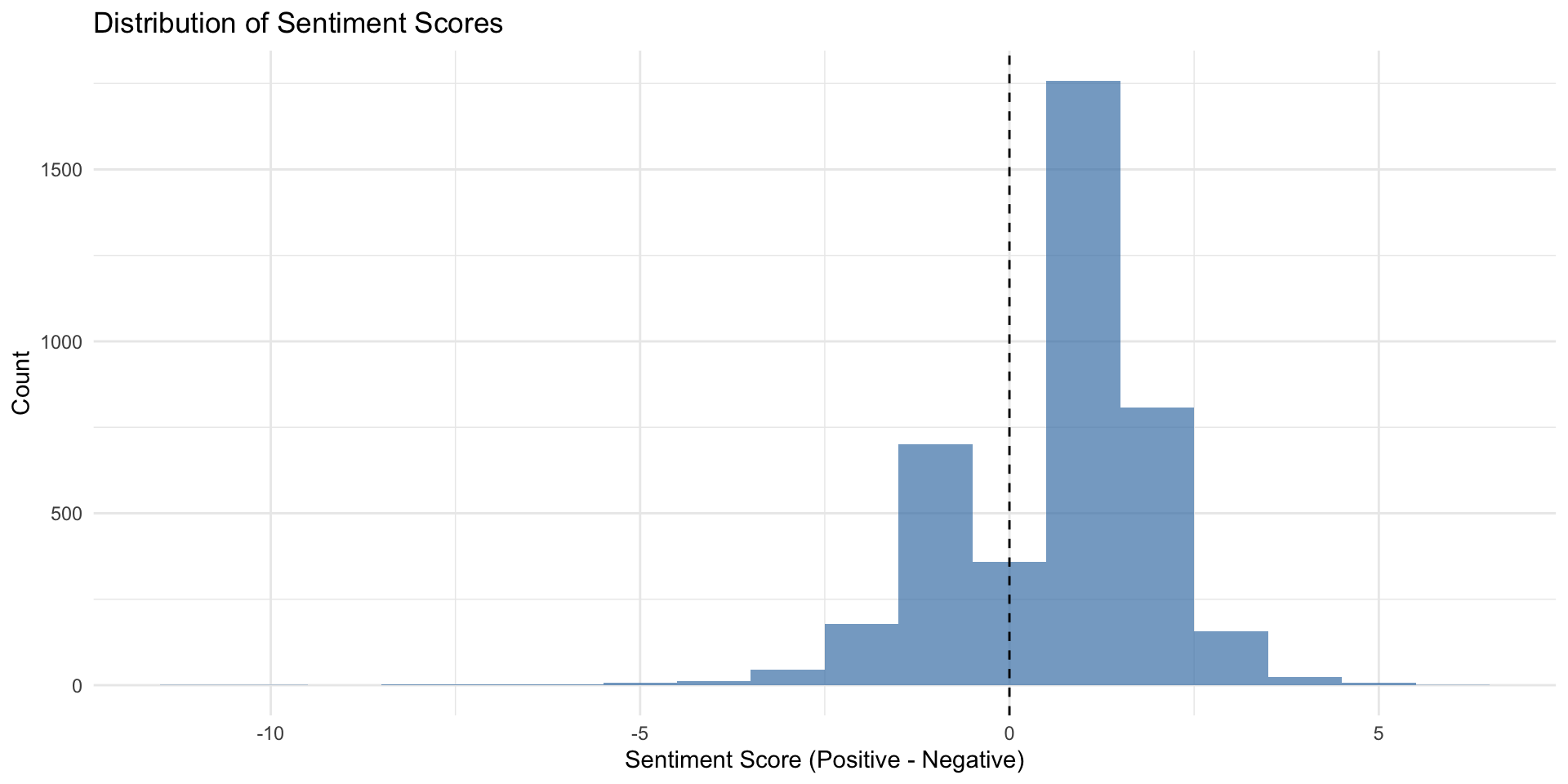
Does sentiment change over time?
Code
sentiment_df %>%
ggplot(aes(x = StartDate)) +
geom_smooth(aes(y = positive, color = "positive sentiment")) +
geom_smooth(aes(y = negative, color = "negative sentiment")) +
scale_color_manual(values = c("#1B4965", "#FF9F1C")) +
labs(
x = "Time (2020)",
y = "Sentiment score"
) +
theme_minimal() +
theme(legend.position = "top")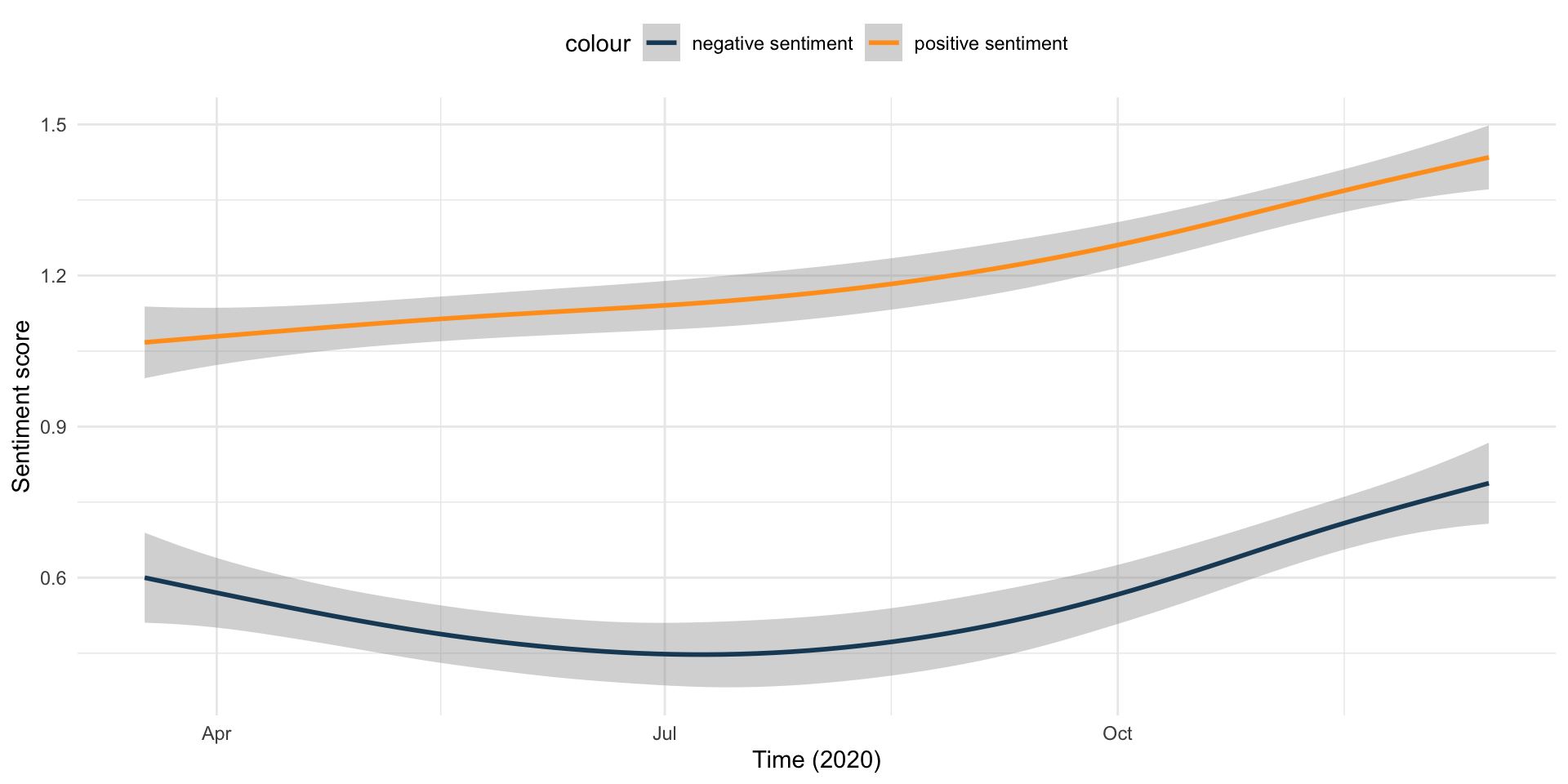
Key Findings
- Most common words reveal attitudes about vaccine safety and effectiveness
- Common bigrams show personal experiences (“already got”, “fully vaccinated”)
- Sentiment analysis reveals mixed but generally positive attitudes
Resources for Learning More
- tidytext documentation: https://www.tidytextmining.com/
- Text Mining with R (free online book)
- UPenn Library
- Michael Clark workshop